This report examines how modern AI methods transform TI by integrating multi-omics datasets, knowledge graphs, machine learning and deep learning architectures, and LLM-based and agentic systems.
Why the choice of the right target determines success in drug discovery
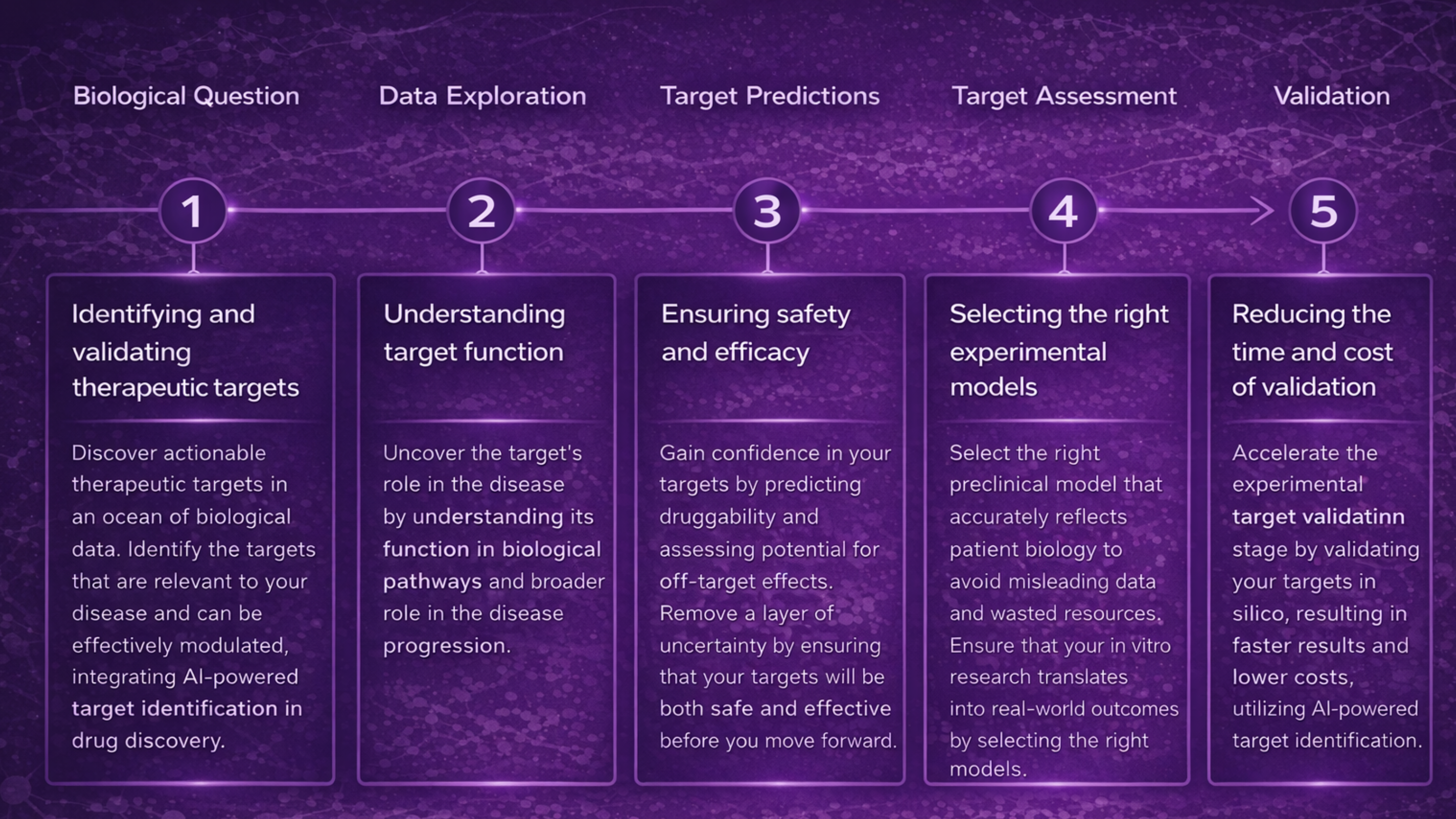
Target selection is the first and fundamental step of the drug discovery pipeline: it defines the biological mechanism to be modulated and sets the stage for all subsequent stages, including compound design, screening, optimization, and clinical translation.
Key points you need to know:
- Target selection defines the biological hypothesis for therapy and links it to mechanism and phenotype –PMC.
- Incorrect or weakly validated targets increase the risk of late-stage failure (e.g., poor efficacy, toxicity, or side effects in clinical trials) – ScienceDirect.
- Mechanistic grounding of targets reduces attrition and increases the probability that downstream molecules will show predictable effects.
Definition of a “disease-relevant target”
A disease-relevant target is a biological entity that satisfies three criteria:
(A) Causal role in pathophysiology
- It participates in the mechanisms driving the disease process.
- Causal evidence arises from genetic, molecular, or pathway studies demonstrating that modulation of the target changes disease phenotype.
(B) Druggability
- Druggability refers to a target’s ability to bind a therapeutic agent with functional consequences for disease.
- Only a small fraction of human biomolecules meet the criteria: targets must be structurally and biochemically amenable to modulation by drug molecules.
(C) Patient-level effect
- Modulation of the target produces therapeutically relevant outcomes in biological or clinical settings (e.g., changes in biomarkers or disease progression metrics) – PMC.
Scale of the problem: why targets remain a bottleneck
2.1. Expensive and Slow R&D, Driven by Biology Risk
Drug discovery and development remains time-consuming and costly, largely because of biology-centric uncertainty at early stages.

The average time from initial discovery through regulatory approval is 10-15 years;
Total capitalized cost per approved drug is estimated at approximately $2.6 billion;
The clinical success rate from Phase I to approval remains below 10-12%.
Primary driver of attrition:
- Most failures occur in Phase II, where lack of efficacy-often due to insufficient biological validation of the target-is the dominant reason.
- Failed validation of disease relevance at the target level leads to downstream failures after significant investment.
The slow and costly nature of the traditional pipeline highlights a structural bottleneck: uncertainty about candidate targets can prolong or derail entire programs.
2.2. Combinatorial Explosion of Potential Targets
The human genome encodes many potential molecular handles for therapeutic intervention, but only a tiny fraction have been exploited successfully.
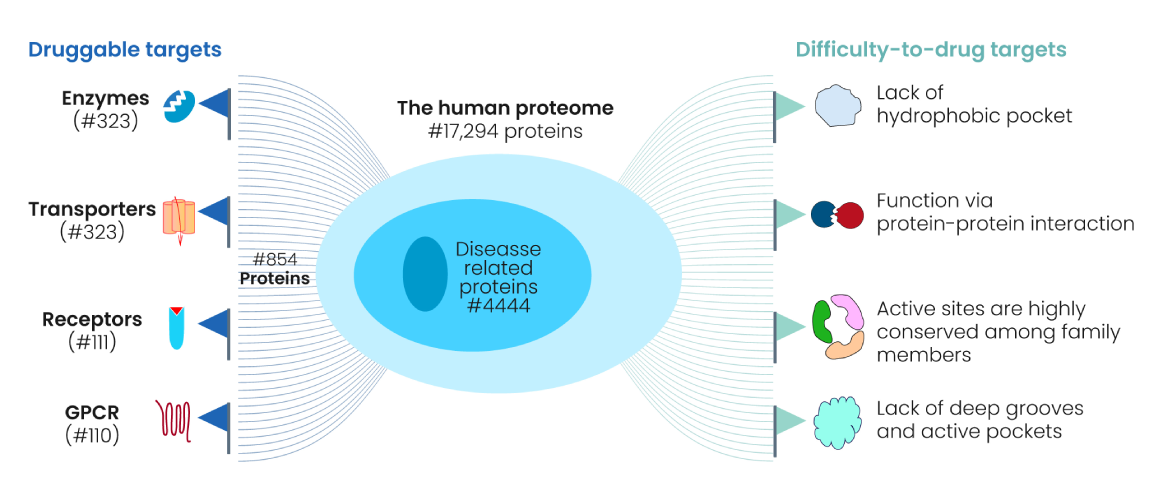
Genomic and proteomic scope:
- Approximately ~20,000 protein-coding genes are expressed in humans – PMC.
- An estimated ~3,000 proteins are considered part of the druggable genome – i.e., amenable to binding by drug-like molecules – NCATS.
- Only a small subset of these have actually been engaged by approved drugs: current FDA-approved drugs target roughly 854 unique proteins (~4 % of the proteome) – The Human Protein Atlas
Interpretation:
- The enormous combinatorial space arising from splice isoforms, post-translational modifications (PTMs), and protein complexes means the number of theoretical potential targets is well over 1 million distinct molecular states.
- Yet >80 % of the potential druggable proteome remains unexplored or insufficiently validated — The Human Protein Atlas.
2.3. Quantifying the Target Bottleneck

2.4. Consequences of the Target Bottleneck
Increased late-stage failures:
- Failed validation of targets leads to poor efficacy in clinical trials, especially Phase II.
- This consumes disproportionate time and resources before stopping expensive programs.
Bias toward well-known biology:
- Research tends to revisit previously studied proteins and pathways, limiting exploration into novel mechanisms.
Long development cycles:
- Lack of integrated target evidence increases cycle time, contributing to the 10-15 year timeline.
Economic inefficiency:
- Upfront biology uncertainty escalates total cost of successful drug development.
AI enables a new perspective on target identification
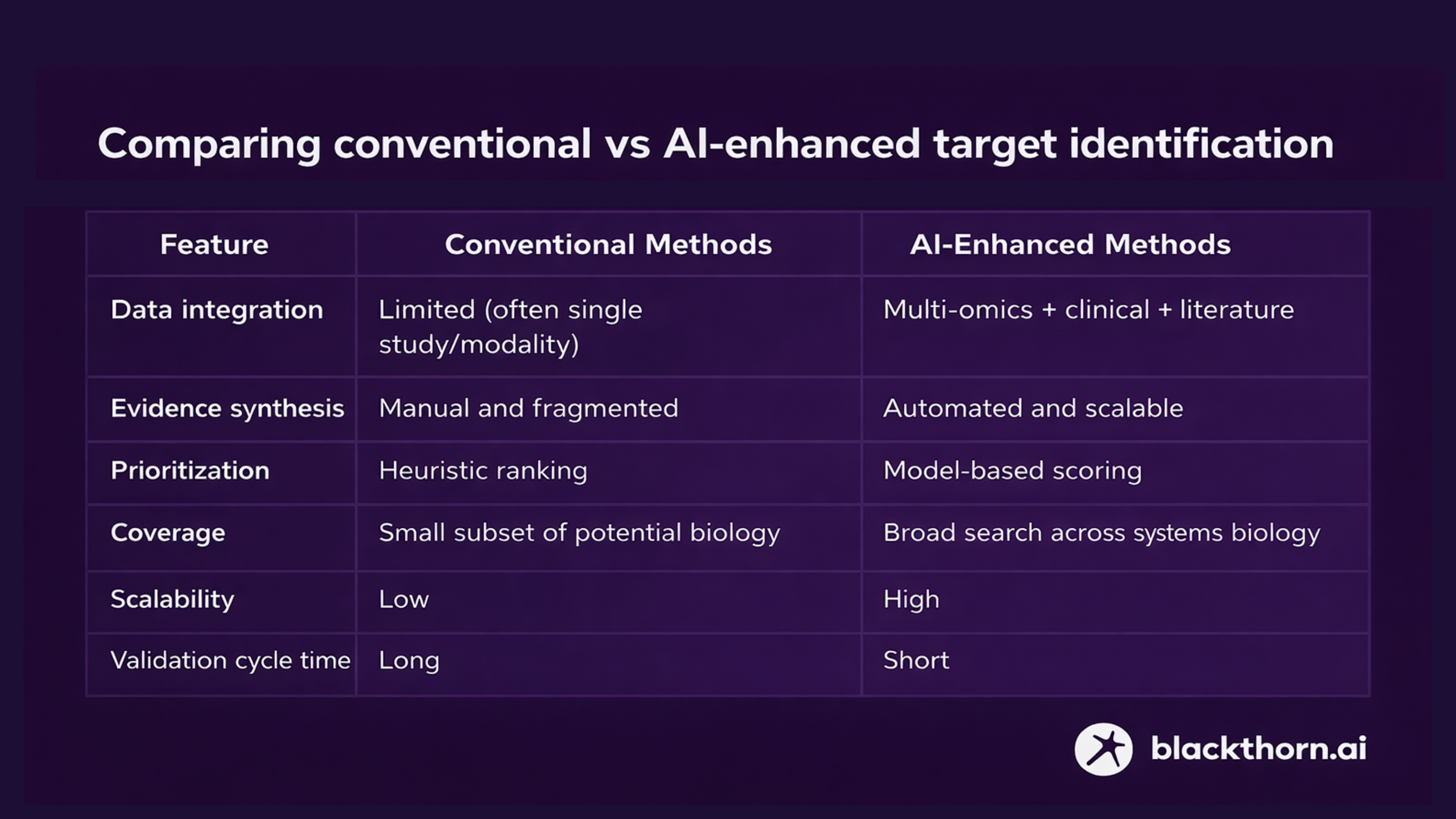
Traditional target identification methods (e.g., single-omics association studies, individual hypothesis testing) are limited by data fragmentation, low scalability, and high false positive rates at scale.
AI-driven approaches change this by:
- Integrating multi-omics datasets: AI + vector databases and/or knowledge graphs can integrate genomics, transcriptomics, proteomics, metabolomics, and phenotypic data into unified representations.
- Synthesizing heterogeneous evidence: Machine learning and neural networks can combine molecular interaction networks with clinical phenotypes and pathway data to prioritize targets that show consistent signals across modalities.
- Mining literature and knowledge resources at scale: Natural language processing and retrieval models can identify relevant experimental evidence and mechanistic literature across millions of publications, improving confidence in target-disease associations.
AI-based methods reduce false positives, increase confidence in disease causality, and expand the pool of candidates by discovering previously unrecognized but mechanistically robust targets. Less routine, time- and resource-consuming lab work. We are talking about months and years.
Comparing conventional vs AI-enhanced target identification
 Ivan Izonin, PhD Sr Scientific Advisor, Artificial IntelligenceTurning target identification into a repeatable, data-driven process with AI
Ivan Izonin, PhD Sr Scientific Advisor, Artificial IntelligenceTurning target identification into a repeatable, data-driven process with AIModern target identification requires integrated multi-omics data, structured biological knowledge, and scalable AI-driven evidence synthesis. This is the foundation we focus on: building AI-ready target discovery pipelines that increase confidence early-before costly downstream decisions are made.
Book a Consultation
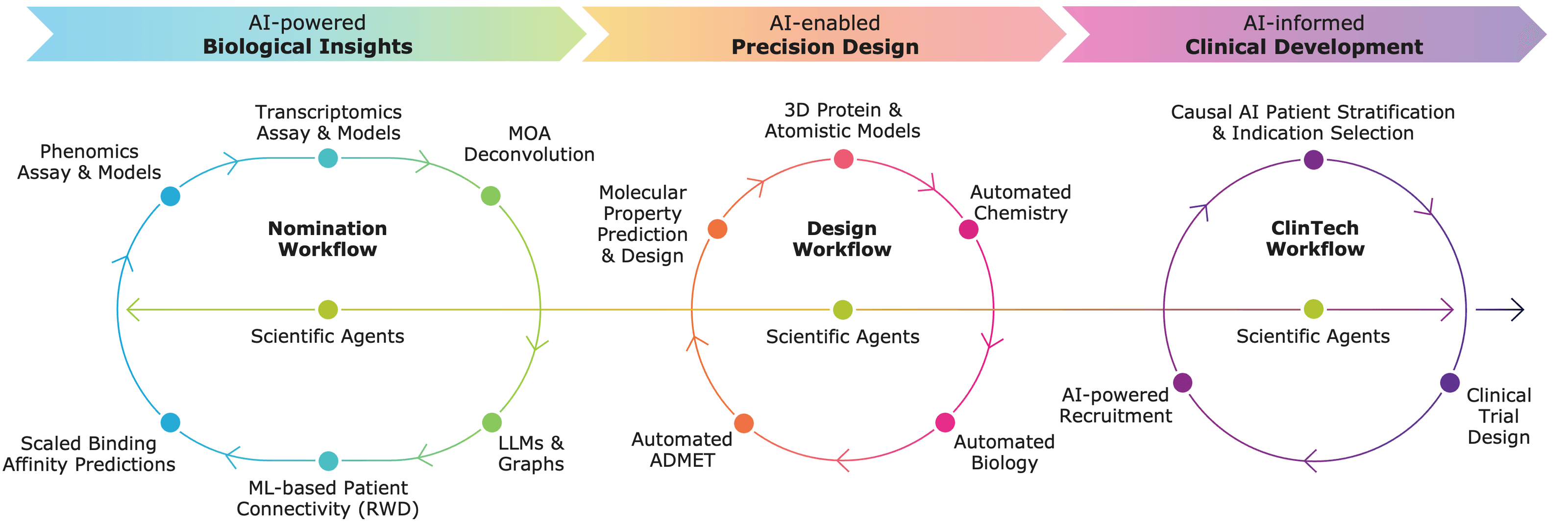
What is an AI-powered target identification?
AI-powered target identification is not a single model but a stack of complementary components that address specific limitations of traditional approaches.
4.1. Multi-omics integration
AI systems integrate genomics, transcriptomics, proteomics, metabolomics, imaging, and clinical data to identify consistent disease signals across biological layers. Multi-omics integration improves resolution of disease mechanisms and reduces false-positive targets from single-modality analyses. Concordant signals across omics layers are stronger indicators of disease causality.
Building such systems requires AI-ready data foundations, multi-omics integration, and workflows designed for scientific decision-making.
Check how these principles are applied
4.2. ML models for target prioritization
Machine learning and deep learning models are used to rank candidate targets based on:
- multi-omics features,
- pathway context,
- disease-gene associations,
- predicted therapeutic impact.
Common model classes:
- supervised models,
- graph neural networks (GNNs),
- autoencoders,
- causal inference models.
These approaches outperform heuristic scoring by 20-40% in target prioritization benchmarks, depending on disease area.
One such implementation is illustrated in a CNS-focused therapeutic discovery platform.
Check how these principles are applied4.3. Knowledge graphs and graph-RAG
Knowledge graphs integrate:
- protein-protein interactions (PPI),
- pathways,
- disease ontologies,
- literature and clinical evidence.
Graph-based reasoning enables:
- identification of indirect disease mechanisms,
- prioritization of targets based on network centrality and causal proximity,
reduction of bias toward overstudied proteins.
AI-assisted literature screening systems construct dynamic evidence graphs that connect genes, diseases, pathways, and publications-enabling target prioritization based on network context rather than publication frequency.
Check how these principles are applied4.4. LLMs and AI agents
Large language models and agentic systems are applied to:
- automated literature review and evidence extraction,
- synthesis of experimental findings across thousands of publications,
- orchestration of iterative hypothesis testing.
LLM-assisted evidence synthesis reduces manual review time by 60-80% while maintaining expert-level recall.
4.5. AI today vs. AI tomorrow in target identification
AI systems:
- propose candidate targets,
- rank them by probability of disease relevance,
- Suggest experimental validation strategies.
4.6. Tomorrow: AI as an agent
Next-generation systems move beyond recommendation toward agentic AI, capable of:
- simulating experimental outcomes,
- iteratively generating and refining hypotheses,
- operating in closed loops with lab automation and digital twins.
Agentic AI frameworks enable continuous learning cycles rather than static scoring.
Practice: real-world cases of AI-powered target identification
5.1 Oncology
Oncology has become the primary proving ground for AI-powered target identification due to the availability of large-scale multi-omics, phenotypic, and clinical datasets.
Industry examples:
Insilico Medicine applies multi-omics integration and AI-driven hypothesis generation to identify novel oncology targets and pathways, combining transcriptomics, proteomics, and phenotypic screening.
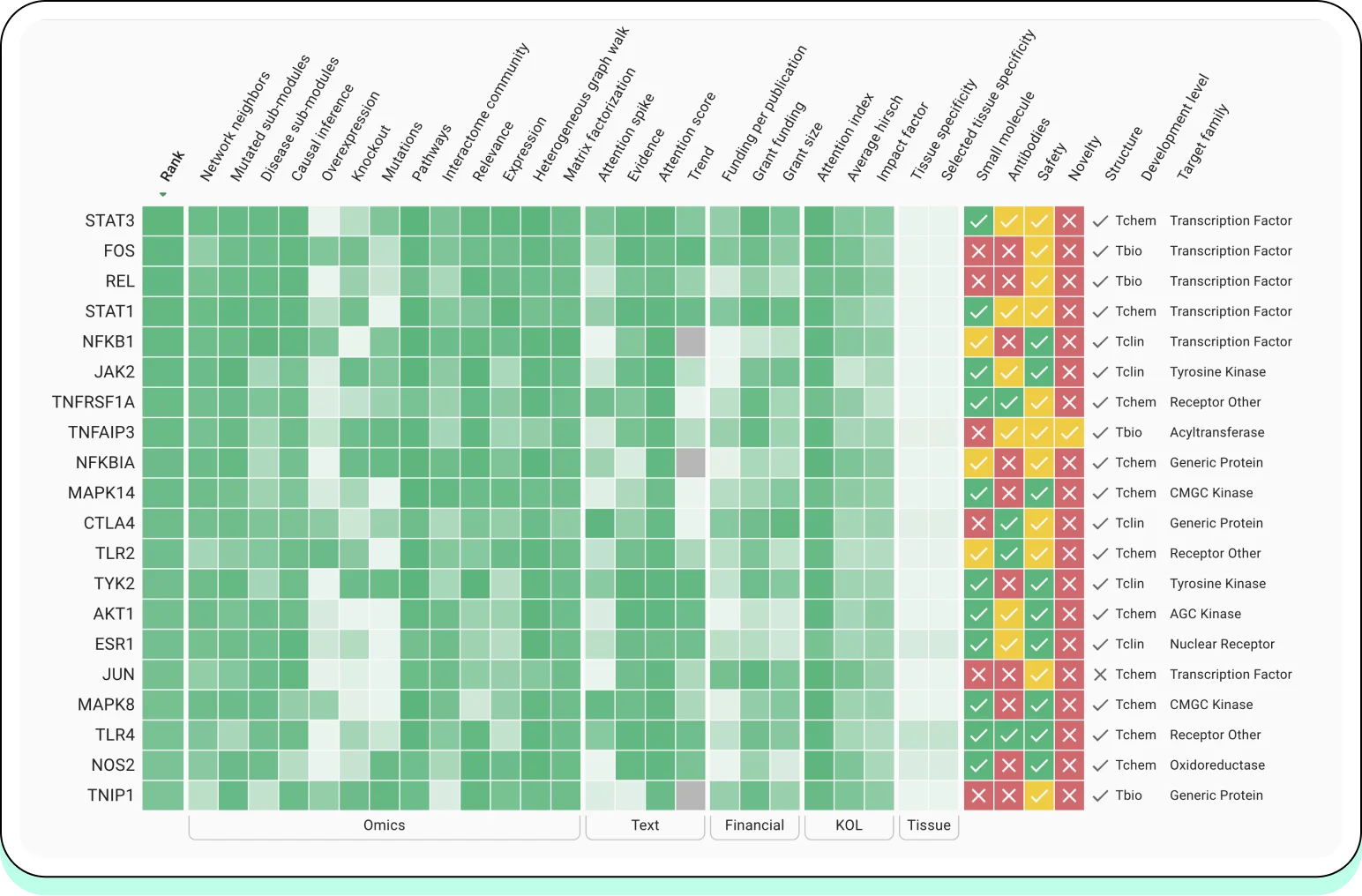
Recursion Pharmaceuticals uses large-scale phenotypic screening coupled with multi-modal data (cell imaging + omics) to infer disease mechanisms and prioritize targets across oncology indications.
Insitro integrates human genetics, functional genomics, and machine learning to identify disease-causal targets, with oncology as a key therapeutic area.
Across these platforms, AI is used not to replace biological reasoning, but to systematically prioritize targets supported by convergent signals across omics layers and phenotypic readouts.
5.2 Alzheimer’s
University of Cambridge + Insilico Medicine demonstrated AI-driven identification of novel disease-relevant targets using integrated omics and network-based modeling, including applications in neurodegenerative diseases such as Alzheimer’s.
This collaboration illustrates how academic biological depth combined with industrial AI infrastructure can surface targets that are difficult to identify using single-modality approaches.
5.3 Fibrosis and IPF
Idiopathic pulmonary fibrosis (IPF) is characterized by complex, multi-cellular pathology and limited therapeutic options, making it a suitable case for AI-driven target discovery.
TherapeutAix identified TNIK as a novel anti-fibrotic target by integrating:
- multi-omics patient data,
- network and pathway analysis,
- large-scale literature mining.
This approach enabled prioritization of TNIK based on pathway-level relevance rather than isolated gene associations, addressing a key limitation of traditional target discovery. The case demonstrates how AI-powered target identification can operate effectively in diseases where mechanisms are distributed across pathways and cell types, rather than driven by single dominant genes.
5.4 Rare diseases
Rare diseases present a fundamentally different challenge for target identification: small cohorts, sparse data, and heterogeneous phenotypes.
Recent reviews highlight how AI enables:
- integration of fragmented genetic and clinical datasets,
- identification of disease-driving biomarkers and targets,
- patient stratification for more precise therapeutic strategies.
AI models can infer disease mechanisms by combining:
- limited omics data,
- prior biological knowledge,
- cross-disease transfer learning.
In this context, AI-powered target identification is less about scale and more about maximizing signal extraction from sparse evidence, making it particularly valuable for orphan indications.
How to launch AI-powered target identification: a practical roadmap
6.1. Readiness assessment: checklist for biotech / pharma / CRO
Before deploying AI-powered target identification, organizations should assess readiness across four dimensions: data, people, infrastructure, and governance.


Deploying AI-powered target identification in practice requires aligning data foundations, ML infrastructure, domain expertise, and governance into a single operational system. For organizations that prefer not to build and integrate this stack in isolation, working with a specialized AI-biotech partner can significantly reduce implementation risk and time to impact.
Learn more6.2. Phased Implementation Roadmap
AI-powered target identification should be deployed incrementally. Attempting full automation from day one increases the risk of failure.
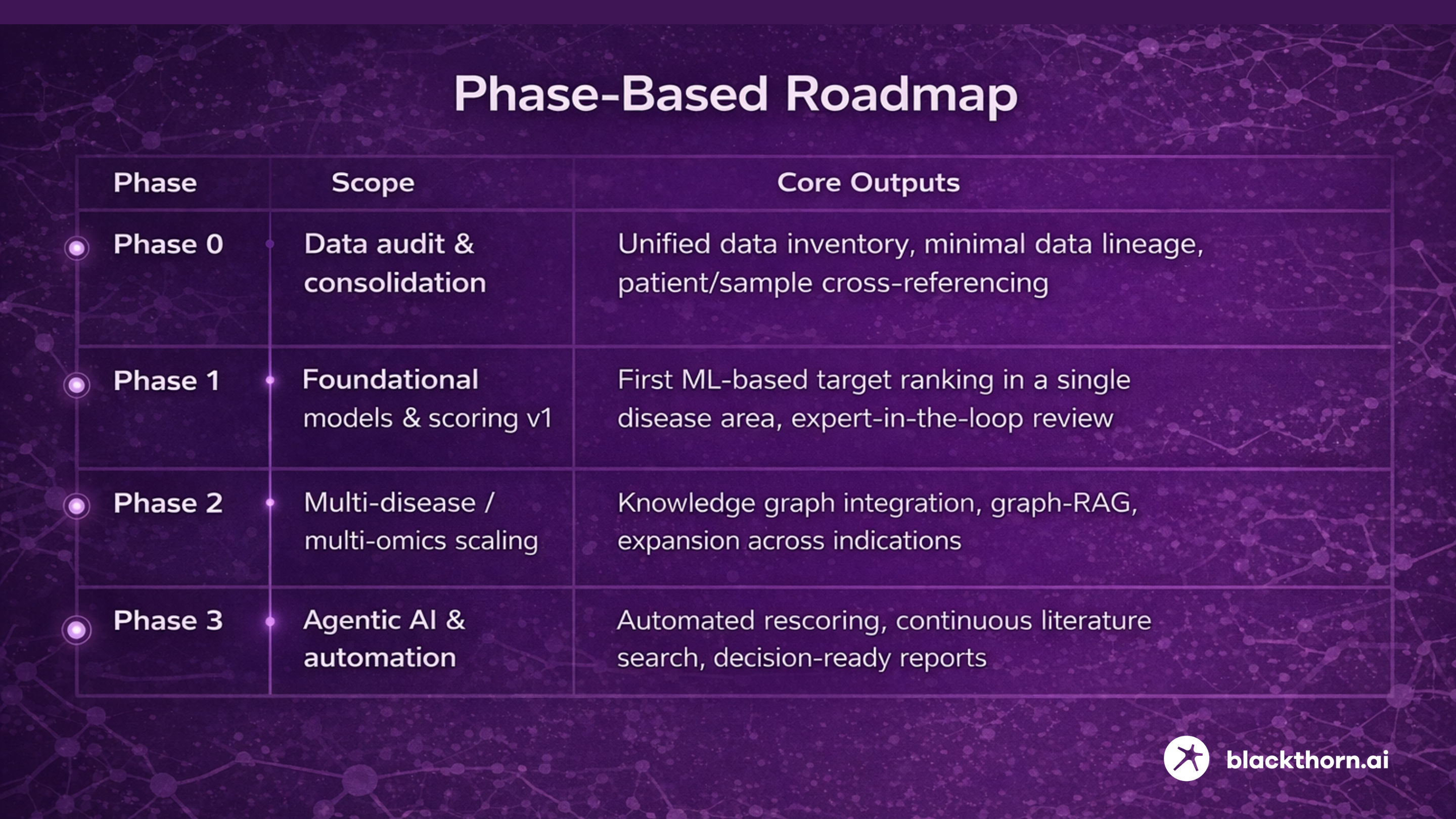
Phase 0 – Data Audit & Consolidation
Objectives:
- Inventory all available omics, clinical, and imaging datasets.
- Identify inconsistencies in identifiers, metadata, and formats.
- Establish minimal data lineage and patient/sample cross-referencing.
Why this matters: poor data integration accounts for 30-40% of downstream model performance loss in biomedical ML systems.
Phase 1 – Foundational Models & Target Scoring (v1)
Objectives:
- Train initial ML models to prioritize targets within a single disease area.
- Combine multi-omics features with pathway and disease-gene associations.
- Integrate human expert review to validate and refine outputs.
Early versions should optimize decision support, not automation. Human-in-the-loop systems reduce false positives by 20-30% compared to fully automated pipelines.
Phase 2 – Multi-Disease and Multi-Omics Scaling
Objectives:
- Extend the platform across multiple therapeutic areas.
- Add knowledge graphs linking genes, pathways, diseases, and literature.
- Introduce graph-RAG for scalable evidence synthesis.
Observed impact: knowledge-graph-based target prioritization improves discovery of non-obvious targets and reduces bias toward well-studied proteins.
Phase 3 – Agentic AI & Automation
AI agents automatically:
- Re-score targets as new omics or clinical data arrive,
- Trigger continuous literature and patent searches,
- Generate structured reports for target review boards.
Why this phase matters: closed-loop AI systems reduce decision latency and support continuous hypothesis refinement.
6.3. KPI and ROI: Measuring Impact of AI-Powered Target ID
To evaluate whether AI truly improves target identification, metrics must focus on decision quality and operational efficiency, not model accuracy alone.
| Metric | What It Measures | Why It Matters |
| Time from raw data to ranked target list | End-to-end cycle time | Faster cycles enable more hypotheses per year |
| Target progression rate | Computational hit → experimental validation → portfolio inclusion | Direct proxy for biological relevance |
| Reduction in late-stage failures | Retrospective analysis of attrition | Late failures are the most expensive |
| Human hours saved | Literature review + data wrangling | Frees scientists for hypothesis design |
| Budget saved | Reduction in early-stage R&D costs through improved target prioritization, automation of literature review, and earlier termination of low-confidence programs | Early-stage biology risk drives the majority of downstream R&D waste; cost savings compound across the pipeline |
Conclusion
AI-powered target identification is fundamentally reshaping where the primary bottleneck in drug discovery lies.
As algorithmic methods mature, the limiting factor is no longer the ability to generate target hypotheses, but the ability to assemble sufficiently high-quality, well-integrated biological data that supports reliable causal inference and clinical relevance.
By systematically integrating multi-omics data, biological networks, and large-scale literature evidence, AI-powered target identification increases the probability that selected targets are truly disease-relevant, mechanistically grounded, and clinically actionable. This shift enables exploration of the largely untapped “dark” portion of the druggable genome, moving beyond historically overstudied proteins and pathways toward novel therapeutic opportunities.
Importantly, this transition is not limited to organizations with massive internal R&D resources.
Startups can apply AI-powered target identification to focus scarce experimental capacity on higher-confidence hypotheses, while large pharma organizations can use it to reduce late-stage attrition, shorten decision cycles, and scale discovery across multiple disease areas.
Realizing these benefits, however, requires more than isolated models-it demands AI-ready data foundations, robust ML infrastructure, and workflows designed for scientific decision-making.

