Imagine a world where machines help decode the mysteries of human biology faster than a lab tech can pipette a sample. That’s the reality artificial intelligence (AI) is carving out in biomedical engineering today. From crunching terabytes of genomic data to spotting tumors in MRI scans that even seasoned radiologists might miss, AI is becoming the Swiss Army knife of modern medicine. In biomedical research, artificial intelligence algorithms sift through decades of protein interaction studies to predict new drug targets – like having a supercharged research assistant who never sleeps.
For diagnosis, tools like LLMs (Large Language Models) digest patient histories and medical literature to suggest rare disease hypotheses, acting as a second opinion when doctors hit a wall. And let’s not forget treatment: AI-powered prosthetics learn users’ movement patterns, adjusting grip strength in real time.
But here’s the kicker: these systems are only as good as the data they’re fed. That’s where knowledge graphs come into play, weaving fragmented biomedical data into structured webs that LLMs can navigate without getting lost in the noise. Think of it as giving a librarian a map instead of tossing them into a book tornado.
This concept comes to life in our Multi-Omics Data Warehouse case, where we designed a scalable infrastructure to integrate genomics, proteomics, and clinical datasets – enabling faster biomedical analysis and AI-driven discovery.
Read the Full CaseApplications of AI in Biomedical Engineering
Let’s roll up our sleeves and dig into where AI’s making waves in biomedical research.
Take medical imaging: convolutional neural networks (CNNs) like U-Net are turning raw MRI slices into 3D tumor maps, pixel by pixel. At Mass General, a model reduced false positives in lung nodule detection by 23% by cross-referencing CT scans with pathology reports stored in a knowledge graph.
Over in genetics, deep learning models predict gene-disease associations by parsing massive biobanks like UK Biobank. For example, DeepVariant slashes errors in identifying insertions/deletions – critical for personalized cancer therapies. Pharma’s getting a facelift too. Generative adversarial networks (GANs) design molecules with specific binding affinities; Insilico Medicine used this to cook up a fibrosis drug candidate in under 18 months. But it’s not all pipettes and petri dishes. Wearable ECG monitors armed with TinyML algorithms now flag atrial fibrillation episodes in real time, sending alerts to cardiologists before a patient feels a flutter. And LLMs? They’re the new lab partners. When a researcher queries, “What’s the link between mTOR pathways and Alzheimer’s?” systems like BioBERT pull answers from 30 million PubMed abstracts – but only if they’re grounded by knowledge graphs to dodge hallucinated references.
In biomedical engineering, AI is also transforming biomedical technology like implantable devices and robotic surgery tools. For instance, AI-driven surgical robots can now perform precise movements that surpass human steadiness, reducing recovery times. Meanwhile, LLMs are helping engineers design better biomedical technology by analyzing decades of research papers and patents, ensuring innovations built on the latest findings.
Challenges and Ethical Considerations in AI for Biomedical Engineering
AI in medicine isn’t all sunshine and rainbows. Let’s start with privacy: training a model on hospital data might leak PHI (Protected Health Information) even with anonymization. A 2022 study showed that 87% of “de-identified” chest X-rays could be re-identified using metadata alone – yikes. Then there’s the elephant in the room: bias. If an artificial intelligence diagnosing skin cancer is trained mostly on light-skinned patients, its accuracy plummets for darker tones. Case in point: a 2021 Nature paper found a 34% drop in melanoma detection accuracy for Black patients compared to White ones. How do we fix this? Some teams are turning to federated learning, where models train across hospitals without sharing raw data. But even that’s no silver bullet; biases in local datasets can still creep in.
And what about errors? An AI recommending the wrong chemo dose could be catastrophic. Adversarial attacks are another headache – adding invisible noise to a retinal scan might trick an artificial intelligence into missing diabetic retinopathy. To build trust, explainability tools like SHAP values are being baked into models, letting doctors see why an AI flagged a tumor as malignant. But until these systems can say, “I’m not sure” instead of guessing, their adoption will hit speed bumps.
In biomedical research, these challenges are even more pronounced. For example, LLMs trained on outdated or biased datasets might generate misleading conclusions, undermining the credibility of biomedical engineering advancements. Addressing these issues requires a mix of better data governance, diverse training datasets, and robust validation frameworks.
How are LLMs used in Biomedicine?
Large language models (LLMs) are advanced AI systems capable of understanding, generating, and manipulating human language by analyzing vast amounts of text.
In biomedicine, LLMs are utilized for a variety of natural language processing (NLP) tasks:
- Text classification
- Information extraction from large texts
- Question answering
- Content creation
- Text summarization
- Machine translation
LLMs are transforming the biomedical field by enhancing drug discovery, streamlining research processes, and accelerating medical research. LLMs offer strategic benefits for the biomedical domain:
- Drug Discovery: Accelerating the identification of potential drug candidates by analyzing scientific literature and existing data speeding up the development process.
- Clinical Research: Enhancing the design and execution of clinical trials by efficiently analyzing and summarizing clinical reports and other relevant research data.
- Enhanced Training: Assisting in the training of new biomedical workers, helping in onboarding.
- Operational Efficiency: Automating routine tasks like data extraction, literature reviews, and report generation, allows researchers to focus on high-impact activities.
While LLM has incredible transformative power, they can’t know all the information, especially in complex domains like biomedicine. They may provide misleading or outdated responses. So it is a good idea to ground LLM using concrete bits of knowledge through Knowledge Graphs.

Discover how Knowledge Graphs can enhance the reasoning capabilities of LLMs in biomedical research and product development. Book a consultation with Ivan Izonin, PhD, Scientific Advisor in Artificial Intelligence at Blackthorn AI, to explore how these technologies can be effectively integrated into your biomedical workflows.
Book a ConsultationThe Power of Knowledge Graphs
Knowledge graphs (KGs) are increasingly used to store semantic relationships and domain knowledge, enabling higher-level reasoning and insights.
Graphs in computer science represent data as nodes (entities) and edges (relationships between entities). Both nodes and edges can have properties. For example, consider representing Genes and Drugs as nodes:
- Gene Node Properties: Name, HGNC symbol, UniProt and Ensembl identifiers, FASTA sequence.
- Drug Node Properties: Name, CheMBL ID, research and approval status, SMILES, commercial names.
- Drug-Gene Relationship: indicates that a drug affects a gene with properties describing the type of interaction and coefficients like IC50, Kd, and KI.
Knowledge Graphs’ strong points
- Integration of Diverse Data Sources: Knowledge graphs can integrate data from multiple sources creating a unified knowledge base.
- Structured Representation: Knowledge graphs represent data in structured form saving relationships between different entities.
- Efficient Querying: Advanced querying capabilities, such as those provided by Cypher query language in Neo4j, enable users to perform complex searches and extract meaningful insights from interconnected data efficiently.
How to integrate Knowledge Graph with LLM?
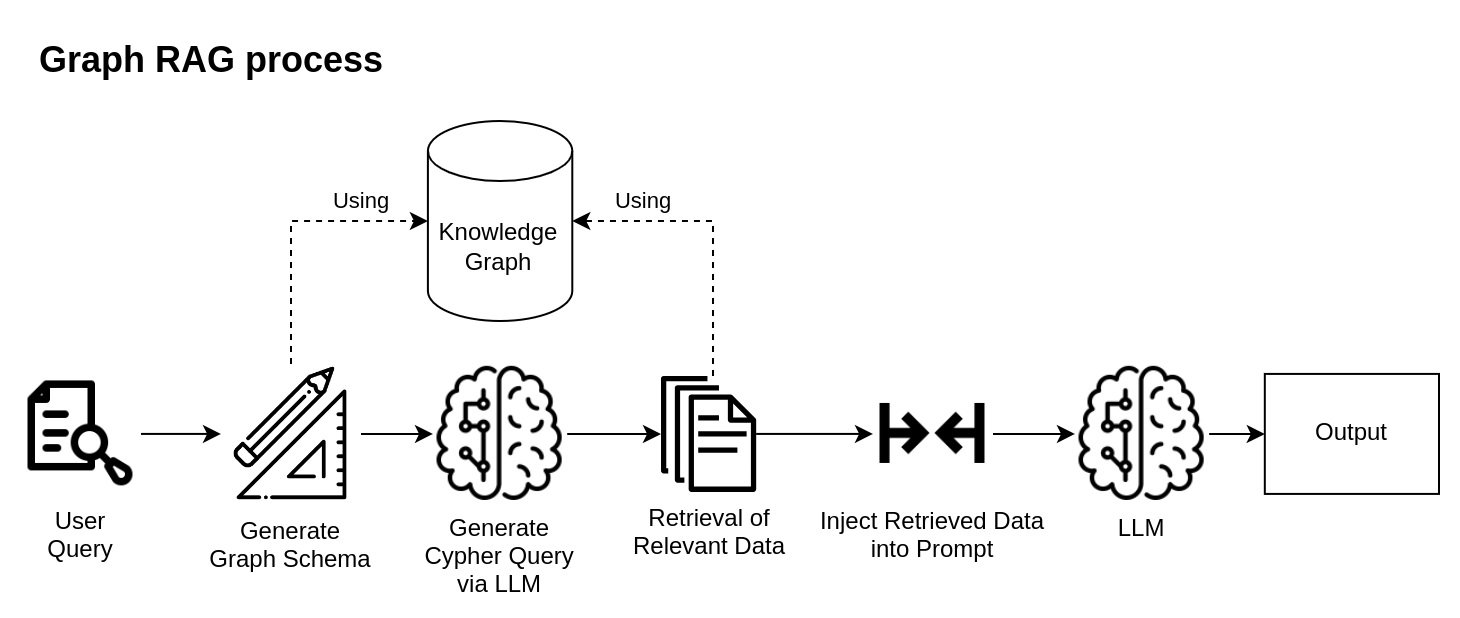
While LLMs offer promising opportunities in the biomedical domain, they often face challenges such as inaccuracy, hallucinations, and a lack of explainability in their responses. Retrieval-augmented generation (RAG) is quickly becoming the industry standard for addressing these issues. The key idea of RAG is to provide LLM with the specific data required to address user questions accurately. However, implementing RAG effectively necessitates a comprehensive and robust knowledge base. You can learn about different RAG approaches in our previous post.
To integrate a Knowledge Graph with an LLM, relevant data must be identified and queried from the graph database. The retrieval results are then injected into the LLM prompt, providing context for the specific question, and allowing the LLM to generate an accurate response.
Case study
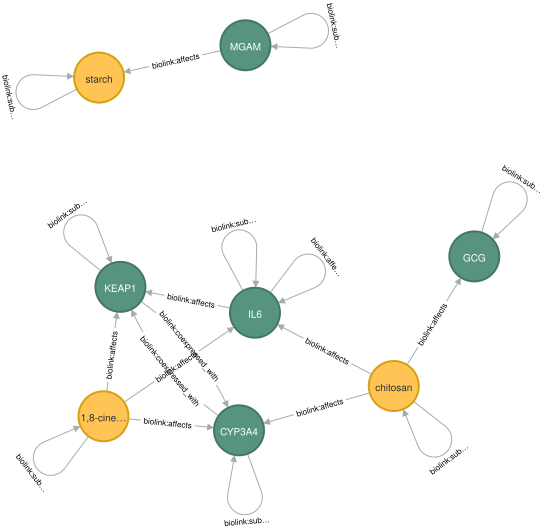
To illustrate Knowledge Graph usage for LLM improvement in the biomedical domain, we used the ROBOKOP knowledge graph. ROBOKOP is an open-source biomedical Knowledge Graph containing millions of relationships derived from numerous biomedical sources, creating a comprehensive network of relationships.
LangChain, a popular framework for LLM-powered application development, provides functionality that supports graph-powered RAG.
First, we need to set up Neo4jGraph. After connecting to the database, the class automatically generates a graph schema by loading all node and edge types along with their properties and identifying possible relationships and constraints. Below is a fragment of such a schema:
Node properties: biolink:PhysicalEssenceOrOccurrent {id: STRING, name: STRING, equivalent_identifiers: LIST, information_content: FLOAT, description: STRING, …} biolink:SequenceVariant {id: STRING, name: STRING, equivalent_identifiers: LIST} biolink:OntologyClass {id: STRING, name: STRING, equivalent_identifiers: LIST, …}\ …
Relationship properties: biolink:subclass_of {biolink:primary_knowledge_source: STRING} biolink:is_nearby_variant_of {biolink:primary_knowledge_source: STRING, distance_to_feature: INTEGER} biolink:coexpressed_with {biolink:primary_knowledge_source: STRING, Coexpression: STRING, Coexpression_transferred: STRING, Experiments: STRING, …} biolink:affects {biolink:primary_knowledge_source: STRING, expressed_in: LIST, p_value: LIST, …}
…
The relationships: (:biolink:PhysicalEssenceOrOccurrent)-[:biolink:affects]->(:biolink:PhysicalEssenceOrOccurrent) (:biolink:PhysicalEssenceOrOccurrent)-[:biolink:affects]→(:biolink:OntologyClass) …
The full ROBOKOP graph schema occupies over 320 thousand tokens (for the GPT-4o tokenizer), exceeding the context window limits of both GPT-4 Turbo and GPT-4o, which is 128 thousand tokens. To address this, we limited the node types to Protein, Gene, Disease, and Drug, and reduced the edge types to the five most important ones. This optimization dramatically decreased the schema token usage to 20.6 thousand tokens, fitting within the context window.
Experiments were conducted using OpenAI API with GPT-4o and GPT-3.5 Turbo models.
Comparison of GPT-4o with and without Integrated KG
For the query “What drugs you know associated with IGF1R receptor?” GPT-4o generates:
Drugs associated with the IGF1R (Insulin-like Growth Factor 1 Receptor) typically target this receptor to inhibit its activity, which can be beneficial in treating certain cancers. Some of the known drugs include:
1. Figitumumab – This is a monoclonal antibody that specifically targets IGF1R.
2. Cixutumumab – Another monoclonal antibody designed to inhibit IGF1R.
3. Dalotuzumab – Also known as MK-0646, this is an IGF1R inhibitor.
4. Ganitumab – This is an investigational drug that targets IGF1R.These drugs are primarily used in research and clinical trials to explore their effectiveness in treating various cancers, as IGF1R is often implicated in tumor growth and survival.
Using Graph RAG, the following information was retrieved from the KG:
[{‘d.name’: ‘PROLINE’, ‘d.id’: ‘CHEMBL.COMPOUND:CHEMBL54922’},
{‘d.name’: ‘Linagliptin’, ‘d.id’: ‘PUBCHEM.COMPOUND:10096344’},
{‘d.name’: ‘Selumetinib’, ‘d.id’: ‘PUBCHEM.COMPOUND:10127622’},
{‘d.name’: ‘Propylene Glycol’, ‘d.id’: ‘PUBCHEM.COMPOUND:1030’},
{‘d.name’: ‘Fulvestrant’, ‘d.id’: ‘PUBCHEM.COMPOUND:104741’},
{‘d.name’: ‘Fulvestrant’, ‘d.id’: ‘PUBCHEM.COMPOUND:104741’},
{‘d.name’: ‘(2R,3S,4R,5R)-2,3,4,5,6-pentahydroxyhexanal’, ‘d.id’: ‘PUBCHEM.COMPOUND:107526’},
{‘d.name’: ‘Nestorone’, ‘d.id’: ‘PUBCHEM.COMPOUND:108059’},
{‘d.name’: ‘Methamphetamine’, ‘d.id’: ‘PUBCHEM.COMPOUND:10836’},
{‘d.name’: ‘Selenious acid’, ‘d.id’: ‘PUBCHEM.COMPOUND:1091’}]
With this context, LLM generates the next answer:
Here are some drugs associated with the IGF1R receptor:
- PROLINE
- Linagliptin
- Selumetinib
- Propylene Glycol
- Fulvestrant
- (2R,3S,4R,5R)-2,3,4,5,6-pentahydroxyhexanal
- Nestorone
- Methamphetamine
- Selenious acid
As shown, the plain LLM provided a list of drugs specifically targeting the IGF1R receptor, including discontinued and current clinical trials, and their commercial names. With RAG, the response included more chemical components affecting the receptor, matching the retrieved information, and ensuring grounded answers, reducing hallucinations.
Let’s extend our question to “What drugs you know associated with IGF1R receptor? Include properties of those associations”. Here is a fragment of plain GPT-4o output:
Several drugs are known to target the IGF1R (Insulin-like Growth Factor 1 Receptor) due to its role in cancer and other diseases. Here are a few notable ones:
1. Figitumumab: This is a monoclonal antibody that specifically targets IGF1R. It works by binding to the receptor and preventing its activation by IGF-1, thereby inhibiting the downstream signaling pathways that promote cell growth and survival. Figitumumab has been investigated in clinical trials for various cancers, including non-small cell lung cancer. …
Graph RAG extracts the following information:
[{‘d’: {‘smiles’: ‘OC(=O)[C@@H]1CCCN1’, ‘CHEBI_ROLE_bacterial_metabolite’: True, ‘description’: ‘Pyrrolidine in which the pro-S hydrogen at position 2 is substituted by a carboxylic acid group. L-Proline is the only one of the twenty DNA-encoded amino acids which has a secondary amino group alpha to the carboxyl group. It is an essential component of collagen and is important for proper functioning of joints and tendons. It also helps maintain and strengthen heart muscles.’, ‘fda_labels’: 21, ‘CHEBI_ROLE_eukaryotic_metabolite’: True, ‘rgb’: 5, ‘CHEBI_ROLE_compatible_osmolytes’: True, ‘sp2_c’: 1, ‘sp3_c’: 4, …}}]
With this context, the LLM generated the following answer:
Here are some drugs associated with the IGF1R receptor and their properties:
1. Proline
– Description: An essential component of collagen, important for joints and tendons.
– Properties: Bacterial, eukaryotic, mammalian, and animal metabolite; osmolyte; micronutrient; drug; pharmaceutical.2. Linagliptin
– Description: Used for type II diabetes treatment.
– Properties: DPP-4 inhibitor, hypoglycemic agent, enzyme inhibitor, pharmaceutical.
…
We can see what happens if RAG can’t find the required information in the knowledge graph for “What diseases are associated with the LTK gene?“. RAG did not retrieve any information, and the model gives the output:
I couldn’t find any specific diseases associated with the LTK gene.
As shown, augmented LLMs provide more concise and grounded answers.
Experiments with Smaller model – GPT-3.5 Turbo
To meet the GPT-3.5-Turbo max context length of 16 thousand tokens we also limited the number of maximal properties in schema per node up to 25. Along with filtering only required node and edge types, this reduced the schema to 1,434 tokens.
This smaller LLM sometimes faced difficulties in generating valid Cypher queries, such as forgetting to escape colons in node and edge types or confusing relationship directions. Additionally, the model occasionally generated queries with intentions slightly different from those requested.
For example, the query “What are the genes associated with multiple sclerosis?” with GPT-4o and its version of KG schema generate Cypher:
MATCH (d:`biolink:Disease` {name: "multiple sclerosis"})<-[:`biolink:causes`]-(g:`biolink:Gene`)
RETURN g;
which retrieve genes that directly cause the disease. However multiple sclerosis disease does not have any relation of type causes with any gene, so with this context model responses:
I currently don’t have access to specific gene information for multiple sclerosis. You might want to consult a medical database or a healthcare professional for detailed information.
The smaller GPT-3.5 Turbo generated a different query, which retrieved any genes that were affected by drugs used to treat multiple sclerosis:
MATCH (d:`biolink:Disease` {name: "multiple sclerosis"})-[:`biolink:treats`]-(drug)-[:`biolink:affects`]-(gene:`biolink:Gene`)
RETURN [gene.name](<http://gene.name/>);
With this query result, GPT-3.5 Turbo generates:
Some of the genes associated with multiple sclerosis are ABL1, MDM4, NLRC4, CNIH4, CXCR2, MDC1, CD28, CDK2, CKS1B, and LGALS9.
This illustrates that the GPT-3.5 Turbo large language model has a misunderstanding of graph topology.
Founded issues of Cypher generation can be solved via prompt tuning, few-shot prompting, query validation, and other methods.
Another common problem discovered for both models is the inability to fetch required nodes from the graph due to the usage of unexpected synonyms. For example, some diseases may have multiple names, but the Cypher queries expect an exact match. This results in missed connections and incomplete data retrieval. To address this issue, robust named entity recognition (NER) and synonym handling mechanisms are required to ensure accurate mapping of terms to their corresponding nodes in the graph.
Conclusions
Integrating Knowledge Graphs with LLMs in the biomedical domain holds significant promise for enhancing the accuracy, relevance, and explainability of AI-generated responses. Using knowledge graphs like ROBOKOP allows the incorporation of diverse knowledge from various biomedical databases within your LLM-powered application.
Benefits
- Cost Optimization: Allow usage of smaller, less specialized LLMs to handle complex biomedical tasks, reducing API usage costs.
- Grounded Answers: Ensures that LLM responses are based on explicit, accurate, and latest knowledge fetched from the knowledge graph, minimizing hallucinations and inaccuracies.
- Improved Relevance: By injecting relevant data into the LLM prompts, the responses are more aligned with the specific questions asked, increasing their utility in complex biomedical domains.
- Increased Explainability: Integrating knowledge graphs helps in tracing the source of information, making AI responses more transparent and trustworthy.
Challenges and Solutions
- Understanding Graph Topology: LLMs may struggle to understand the complex relationships and structures within a knowledge graph. Solution: provide examples in the prompt to help the LLM understand the graph’s structure and relationships.
- Huge Schema Size: The size of the schema can be overwhelming. To make the schema shorter and more targeted to the question, RAG techniques can be used. For example, we could find the required node and edge types using search with question embeddings. Solution: Use embeddings to filter and retrieve only the most relevant parts of the schema, ensuring the LLM context window is efficiently utilized.
- Fetching Required Nodes: This is straightforward for genes due to the usage of standard HGNC identifiers. For diseases and drugs, named entity extraction is necessary to handle synonyms effectively. Solution: Implement robust named entity recognition (NER) techniques to accurately identify and match synonyms, ensuring consistent data retrieval.
If you are interested in Knowledge-Graph-powered LLM applications, feel free to contact us at blackthorn.ai.
In conclusion, integrating Knowledge Graphs with LLMs enhances the capabilities of AI in the biomedical field, leading to improved patient outcomes, more efficient healthcare operations, and accelerated medical research.
Future Trends in AI and Biomedical Engineering
What’s next? Picture digital twins – virtual replicas of patients that simulate how a drug might affect their organs. Companies like Unlearn.ai are already building these using EHR data fused with knowledge graphs, letting clinicians test treatments on a digital clone before prescribing. Neural networks are also getting sharper at predicting diseases. Take Google’s DeepMind: their Graph Neural Network (GNN) predicts acute kidney injury 48 hours before onset by analyzing lab trends and vital signs as interconnected nodes.
Automated research labs are another frontier. Imagine AI-driven robots conducting high-throughput experiments; Carnegie Mellon’s “Genesis” platform designs and runs CRISPR trials autonomously, tweaking parameters based on real-time results. And let’s not forget LLMs. Future models might draft entire research papers, but only if anchored by knowledge graphs that tether claims to verified sources. The catch? These systems will need real-time updates – like integrating the latest preprint findings on SARS-CoV-2 variants without retraining from scratch.
In biomedical engineering, the integration of artificial intelligence and biomedical technology is set to redefine patient care. For instance, AI-powered wearable devices could continuously monitor health metrics, predicting heart attacks or strokes before they happen. Meanwhile, LLMs could revolutionize how we approach biomedical research, synthesizing insights from millions of studies to accelerate discoveries. It’s a high bar, but hey, nobody said curing diseases would be a walk in the park.